On the structure of things.
Notes on the WorkflowR package: A streamlined process to code, document, build figures and share analysis.

To be more specific, on the structure of a data-analysis folder 📂. This small note describes generating and using an all-in-one structured project folder with data, scripts, documentation, and a sharable HTML page.
For every experiment I do, I like to explore data, analyse, create figures, and add notes 📝 to figures, all leading to, hopefully, some observation or conclusion. Ideally, I would share and discuss findings, after which I would need to find back the code to re-explore or edit the figures.
After several years of manually creating some structure of folders with figures, occasionally a ppt summary or Rmd files, I figured a function should create a template folder structure. Two years ago, I found exactly that and more!
An R package 📦 made J. Blishak for reproducible and sharable data science.
WorkflowR 📦 has it all. It has a single function to start a project, generating all the basics I need, git version control, the option to create sharable HTML output and all sorts of good-practice coding checks along the way.
Documentation with several tutorials is available at the package website. In brief, one can create a project 📂 with the following structure:
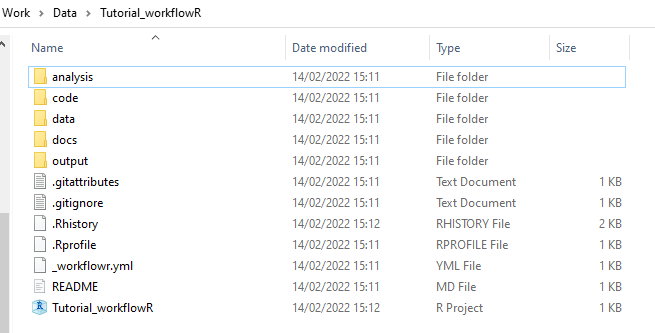
We then create Rmd files within the analysis folder with descriptions around code and figures. We can create an HTML file from a specified Rmd-file with the command build. The HTML file will fit into a linked web page found in the docs folder (depending on your configuration or version). Put raw and metafiles in the data folder to keep everything organised while scripts go into the code folder. The working directory contains an Rproject file, gitignore and other essential documents that can be modified as required.
For my department computational supergroupmeeting, I prepared a quick overview and small example in ppt. All of the information in the ppt comes from the original package documentation. The generated Tutorial_workflowR pages are available as GitHub page.

Original manuscript describing the ‘WorkflowR’ package:
Blischak JD, Carbonetto P, and Stephens M. Creating and sharing reproducible research code the workflowr way. F1000Research 2019, 8:1749